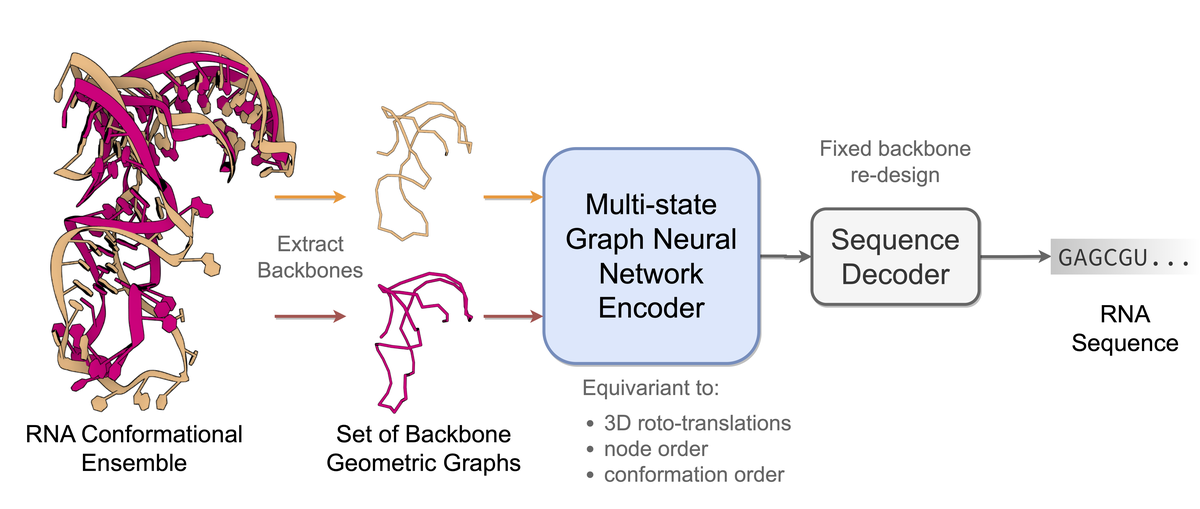
Abstract
Computational RNA design tasks are often posed as inverse problems, where sequences are designed based on adopting a single desired secondary structure without considering 3D geometry and conformational diversity. We introduce gRNAde, a geometric RNA design pipeline operating on 3D RNA backbones to design sequences that explicitly account for structure and dynamics. Under the hood, gRNAde is a multi-state Graph Neural Network that generates candidate RNA sequences conditioned on one or more 3D backbone structures where the identities of the bases are unknown. On a single-state fixed backbone re-design benchmark of 14 RNA structures from the PDB identified by Das et al. [2010], gRNAde obtains higher native sequence recovery rates (54% on average) compared to Rosetta (45% on average), taking under a second to produce designs compared to the reported hours for Rosetta. We further demonstrate the utility of gRNAde on a new benchmark of multi-state design for structurally flexible RNAs, as well as zero-shot ranking of mutational fitness landscapes in a retrospective analysis of a recent RNA polymerase ribozyme structure.
Getting started with 3D RNA modelling as a machine learner
If you’re already an expert in RNA structure modelling and want to get started with using gRNAde, check out the tutorial notebook within this directory. Collab version:
If you’re a computer scientist or machine learner without much background in biology but keen to enter the RNA world, read on…
Why RNA? A personal account.
The Covid-19 pandemic was a very challenging period for me personally. At the same time, I got interested in reading about and understanding some of the science behind the disease (I had last formally studied biology in 10th grade and didn't enjoy all the fact memorisation). In 2022, I began my PhD at Cambridge with Pietro Liò and was interested in biomolecules. I inherently knew that I wanted to motivated my work as a machine learner by biological problems that (hopefully) matter, but I didn't know much biology so I started having conversations with Pietro and my labmates, as well as reading lots of popular science books. I found casual conversations and books far more approachable, unlike textbooks or courses. The pandemic was coming under control largely due to mRNA vaccines and I was feeling very inspired after reading about the scientific lives of Jennifer Doudna (Nobel Prize for CRISPR) and Venki Ramakrishnan (Nobel Prize for Ribosomes), as well as several of Siddhartha Mukherjee's books.One uniting theme across everything I was reading and what was happening around me was the central role played by RNA molecules. Everybody (including myself) in deep learning was very excited about modelling proteins after AlphaFold happened, but a chance conversation with Roger Foo suggested that RNA might be something that’s currently more exciting and novel to biologists. I can’t explain precisely why, but I found something very interesting about RNA’s biochemistry, the intricate structures it folds into, and the stories of the research as well as the scientists behind them. I also found the community to be very welcoming and friendly to newcomers :)
Disclaimer. This document contains a currated list of resources I’ve been using in order to understand RNA biology and feel motivated to work on RNA design. I’m not an expert at all, so it is very likely that I’ve missed something important. None the less, I do hope these resources will be useful for someone like myself a couple years ago!
Before you begin, if you like games, do try and play the introductory parts of Eterna. I can’t think of a better way to start your journey in learning about RNA while having some fun!
🧬 RNA Biochemistry and Structural Biology
I can highly recommend working through the RNA 3D Structure Course by Craig L. Zirbel and Neocles Leontis at Bowling Green State University. These self-contained notes are perfect for learning topic-by-topic at your own pace, and I find myself coming back to them frequently for reference. Here are three videos to go along with the notes:
- Anna Marie Pyle’s excellent introduction to RNA structure for iBiology.
- A similar lecture as part of the MIT course by Barbara Imperiali.
- Eric Westhof’s talk at CASP 15 about what makes RNA structure different from proteins and how to compare RNA structures. You can feel how passionate he is about all the small but important details!
For understanding the broader biological context, I simultaneously listened through MIT 7.016 Introductory Biology, which has really passionate lecturers and starts from the basics.
Thoughts on how to think (and talk) about RNA structure. An approachable yet thorough paper introducing RNA structure. I keep finding myself coming back to refer to some details in this manuscript and highly recommend it.
Lastly, if you want to work through a textbook, I’m sure there are several nice ones out there. I’ve been referencing Principles of Nucleic Acid Structure from time to time because Pietro Liò very kindly gave me his copy.
I’m also enjoying the new RNA Biology Coursera course by Rhiju Das and the Das Lab at Stanford. I would perhaps start here if I were starting from scratch.
🎨 RNA Design (and Biomolecule Design, more broadly)
Geometric Deep Learning for designing protein structure is very exciting at the moment! I would start by watching the latest talk I can find by David Baker (example) and reading/watching talks about the 3 main tools for protein structure modelling and design: AlphaFold2, ProteinMPNN, and RFdiffusion.
- Nazim Bouatta’s lectures on AlphaFold.
- ProteinMPNN by Justas Dauparas.
- RFdiffusion by Joe Watson and David Juergens.
- A new survey which does a fantastic job overviewing these methods as well as protein language models (sequence-centric approach to design).
Rhiju Das’s excellent talk on 3D RNA Modelling and Design poses a question that then captivated me: Can we bring to bear the success of these tools from the world of proteins to RNA?
- The accompanying perspective article.
- RNAMake by Joseph Yesselman introduces a (non-ML) algorithm for aligning RNA motifs like lego blocks. It is particularly interesting to get a grasp of what sort of design scenarios one may be interested in. Joseph’s talk is also very nice.
- I also read through Rhiju’s early works introducing the structure-based design paradigm for RNA; just sort his Google Scholar by date and scroll down. The paper on Rosetta for RNA felt like a very important one, in particular.
Ewan McRae’s talk on RNA origami, another emerging non-ML paradigm for structural RNA design through assembling modular building blocks.
For something a bit different but thought-provoking, Phil Holliger’s talk on evolutionary approaches to designing biomolecules. “Evolution is the most powerful algorithm currently known to man”, and its perhaps worth pondering how structure-based or de-novo design can augment, automate, or complement parts of the already very powerful directed evolution approach to designing biomolecules. Phil also briefly discusses XNAs which are designed nucleic acids with rather profound implications for the origins of life itself; I do encourage you to go down that rabbit hole!
📦 Datasets
- RNASolo, a repository of processed PDB-derived RNA 3D structures. Go ahead and look at some of the RNA structures in the viewer, see if you like how they twist and turn!
- RNA 3D Hub, a repository of RNA structural annotations, motifs, and non-redundant (clustered) sets.
- Introduction to the PDB file format.
📝 More Papers
Coarse-grained modelling of RNA 3D structure, an excellent overview of strategies for representing RNA structures as input for computational pipelines. This paper from Janusz Bujnicki’s Genesilico group (check out their website for lots of great resources) focussed on folding/structure prediction, but coarse-graining is a universal idea applicable across all machine learning tasks for RNA 3D structure.
The roles of structural dynamics in the cellular functions of RNAs and RNA conformational propensities determine cellular activity, two important papers advancing a growing understanding of how RNAs (and other biomolecules for that matter) are not rigid 3D objects but rather a dancing ensemble composed of multiple structural or functional states. We were inspired to build multi-state Graph Neural Networks for gRNAde based on this line of work.
When will RNA get its AlphaFold moment?. Based on the results at the latest CASP, perhaps not till we improve our training datasets (analysis video by Rhiju Das).
Some interesting surveys:
My playlist with most of the videos references in this document.