Abstract
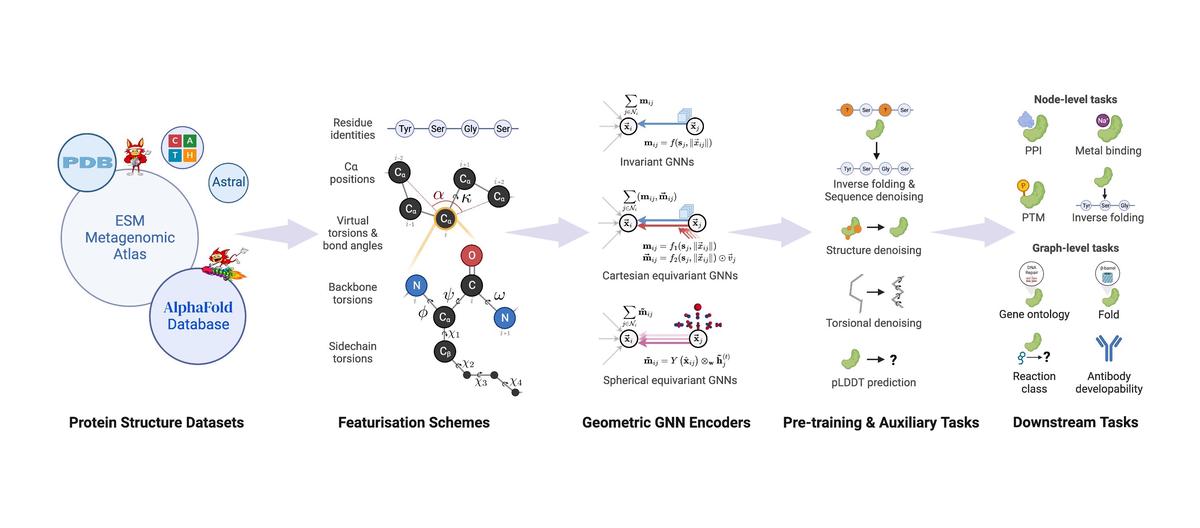
We introduce ProteinWorkshop, a comprehensive benchmark suite for evaluating protein structure representation learning methods. We provide large-scale pre-training and downstream tasks comprised of both experimental and predicted structures, offering a balanced challenge to representation learning algorithms. We benchmark state-of-the-art protein-specific and generic geometric Graph Neural Networks and the extent to which they benefit from different types of pre-training. We find that: (1) Pretraining and auxiliary tasks consistently improve the performance of both rotation-invariant and equivariant geometric models; (2) More expressive equivariant models benefit from pretraining to a greater extent compared to invariant models; (3) Our open-source codebase reduces the barrier to entry for working with large protein structure datasets by providing storage-efficient dataloaders from large-scale predicted structures including AlphaFoldDB and ESM Atlas, as well as utilities for constructing new tasks from the entire PDB. ProteinWorkshop is available at: https://github.com/a-r-j/ProteinWorkshop.